Revamped design could take powerful biological computers from the test tube to the cell
Very small biological computers made of DNA could revolutionize the way we diagnose and handle a slew of illnesses, at the time the technologies is fully fleshed out. On the other hand, a significant stumbling block for these DNA-based mostly devices, which can function in both of those cells and liquid solutions, has been how limited-lived they are. Just one use and the pcs are put in.
Now, scientists at the Countrywide Institute of Requirements and Know-how (NIST) might have made extensive-lived biological computer systems that could potentially persist within cells. In a paper revealed in the journal Science Advances, the authors forgo the common DNA-based method, opting as an alternative to use the nucleic acid RNA to develop computers. The success display that the RNA circuits are as reliable and flexible as their DNA-based mostly counterparts. What is additional, living cells may well be able to build these RNA circuits consistently, one thing that is not easily attainable with DNA circuits, further more positioning RNA as a promising prospect for impressive, extended-long lasting organic computer systems.
Substantially like the computer or sensible product you are probably looking through this on, organic personal computers can be programmed to carry out unique kinds of responsibilities.
“The variance is, as an alternative of coding with types and zeroes, you create strings of A, T, C and G, which are the 4 chemical bases that make up DNA,” mentioned Samuel Schaffter, NIST postdoctoral researcher and lead writer of the analyze.
By assembling a distinct sequence of bases into a strand of nucleic acid, scientists can dictate what it binds to. A strand could be engineered to connect to specific bits of DNA, RNA or some proteins connected with a ailment, then set off chemical reactions with other strands in the exact same circuit to system chemical information and finally make some type of useful output.
That output might be a detectable sign that could aid medical diagnostics, or it could be a therapeutic drug to handle a sickness.
Having said that, DNA is not the sturdiest substance and can promptly appear aside in specific ailments. Cells can be hostile environments, given that they generally contain proteins that chop up nucleic acids. And even if DNA sequences adhere all-around prolonged ample to detect their target, the chemical bonds they kind render them worthless afterward.
“They can not do things like consistently keep track of patterns in gene expression. They are one use, which indicates they just give you a snapshot,” Schaffter reported.
Staying a nucleic acid as perfectly, RNA shares lots of of DNA’s woes when it will come to becoming a organic computer system building block. It is prone to rapid degradation, and after a strand chemically binds to a focus on molecule, that strand is completed. But as opposed to DNA, RNA could be a renewable resource in the ideal conditions. To leverage that benefit, Schaffter and his colleagues initial needed to show that RNA circuits, which cells would theoretically be able to make, could function just as very well as the DNA-centered variety.
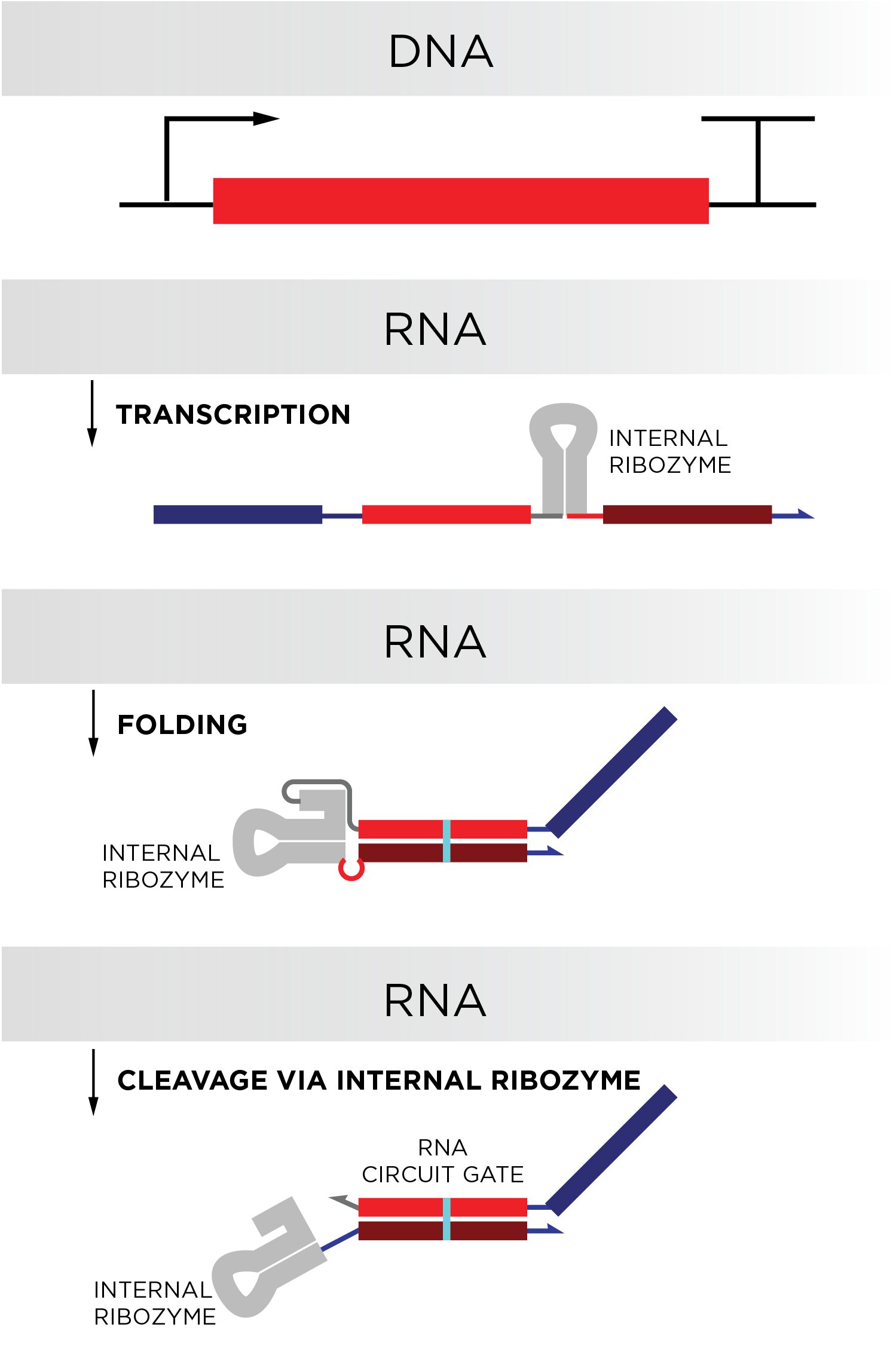
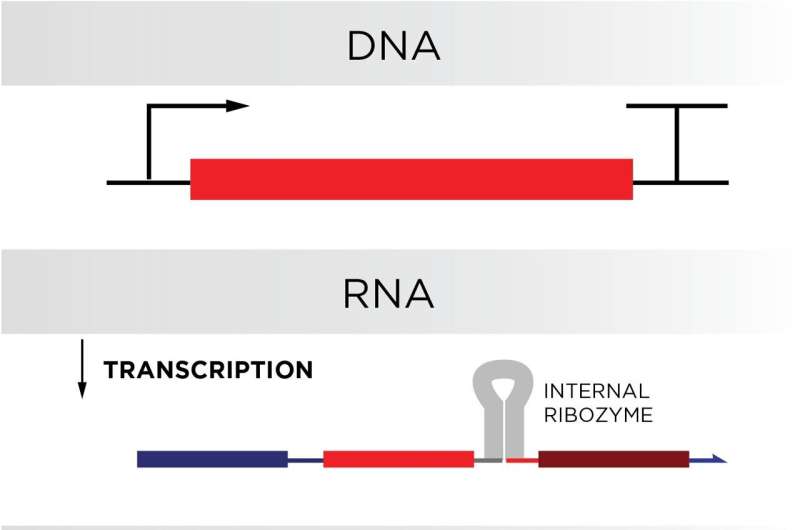
RNA’s edge above DNA stems from a pure mobile system named transcription, wherein proteins make RNA on a continuous basis applying a cell’s DNA as a template. If the DNA in a cell’s genome coded for the circuit components in a organic pc, then the cell would make the laptop factors constantly.
In the organic computing process, single strands of nucleic acids in a circuit can quickly stop up sure to other strands in the identical circuit, an undesired result that helps prevent circuit factors from binding to their intended targets. The structure of these circuits usually indicates that various components will be purely natural matches for just about every other.
To avoid undesired binding, DNA sequences that are part of personal computers recognised as strand displacement circuits are normally synthesized (in machines relatively than cells) separately and in a double-stranded sort. With each and every chemical foundation on each and every strand certain to a foundation on the other, this double strand acts as a locked gate that would only unlock if the focus on sequence came along and took the place of just one of the strands.
Schaffter and Elizabeth Strychalski, leader of NIST’s Cellular Engineering Group and co-writer of the study, sought to mimic this “locked gate” functionality in their RNA circuit, maintaining in thoughts that, in the long run, cells would have to develop these locked gates them selves. To established cells up for good results, the scientists wrote the sequences so that 1 50 percent of the strands could bind flush with the other 50 percent. Binding this way, RNA sequences would fold on on their own like a hotdog bun, making certain they are in a locked point out.
But to work correctly, the gates would want to be two chemically bound but unique strands, far more like a hamburger bun or sandwich than a hotdog bun. The crew obtained the double-stranded structure in their gates by coding in a extend of RNA termed a ribozyme in the vicinity of the folding place of the gates. This distinct ribozyme—taken from the genome of a hepatitis virus—would sever alone right after the RNA strand it was embedded in folded, generating two individual strands.
The authors examined irrespective of whether their circuits could accomplish essential reasonable operations, like only unlocking their gates under unique scenarios, these types of as if just one of two precise RNA sequences was existing or only if the two ended up at the similar time. They also designed and examined circuits created of many gates that performed diverse sensible functions in sequence. Only when these circuits encountered the proper mixture of sequences, their gates would unlock a single by a single like dominoes.
The experiments associated exposing different circuits to items of RNA—some of which, the circuits have been developed to attach to—and measuring the output of the circuits. In this circumstance, the output at the finish of each and every circuit was a fluorescent reporter molecule that would mild up at the time the final gate was unlocked.
The scientists also tracked the rate at which the gates unlocked as the circuits processed inputs and in contrast their measurements to the predictions of pc models.
“For me, these required to operate in a check tube as predictively as DNA computing. The great factor with DNA circuits is most of the time, you can just compose out a sequence on a piece of paper, and it’s going to get the job done the way you want,” Schaffter reported. “The critical thing here is that we did locate the RNA circuits ended up really predictable and programmable, substantially a lot more so than I thought they would be, basically.”
The similarities in functionality among DNA and RNA circuits could suggest that it could be useful to swap to the latter, because RNA can be transcribed to replenish a circuit’s components. And many present DNA circuits that researchers have by now created to accomplish numerous tasks could theoretically be swapped out for RNA variations and behave the identical way. To be confident, although, the authors of the research want to push the engineering further.
In this research, the authors demonstrated that transcribable circuits operate, but they have not manufactured them employing the authentic mobile machinery of transcription nonetheless. Instead, devices synthesized the nucleic acids by way of a course of action very similar to that utilized to generate DNA for investigation. Getting the following stage would call for inserting DNA into the genome of an organism, where by it would serve as a blueprint for RNA circuit parts.
“We’re fascinated in putting these in micro organism following. We want to know: Can we offer circuit types into genetic material utilizing our method? Can we get the exact kind of efficiency and conduct when the circuits are inside cells?” Schaffter claimed. “We have the probable to.”
Experts acquire novel DNA logic circuits
Samuel Schaffter, Co-transcriptionally encoded RNA strand displacement circuits, Science Advancements (2022). DOI: 10.1126/sciadv.abl4354. www.science.org/doi/10.1126/sciadv.abl4354
Quotation:
Revamped style could acquire potent organic personal computers from the check tube to the cell (2022, March 23)
retrieved 24 March 2022
from https://phys.org/news/2022-03-revamped-potent-organic-tube-mobile.html
This document is subject to copyright. Aside from any fair working for the intent of private examine or study, no
part may well be reproduced without the written permission. The content material is supplied for information and facts functions only.
